
Different modes of protein–DNA binding. The profiled protein is shown... | Download Scientific Diagram
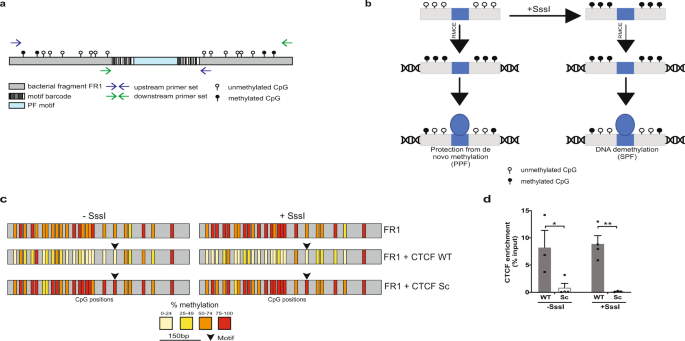
High throughput screening identifies SOX2 as a super pioneer factor that inhibits DNA methylation maintenance at its binding sites | Nature Communications

Identification of a novel class of genomic DNA-binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53
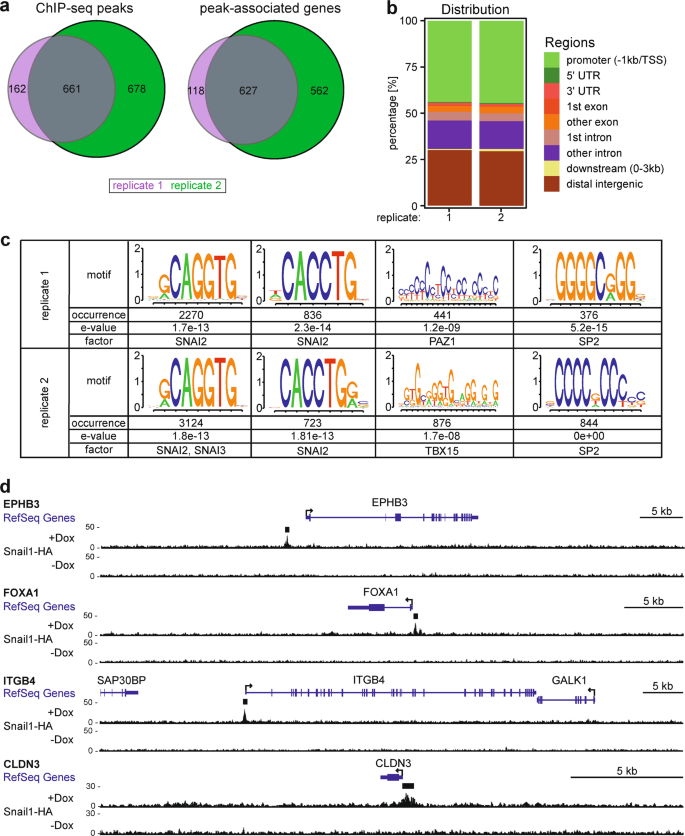
Genome-wide mapping of DNA-binding sites identifies stemness-related genes as directly repressed targets of SNAIL1 in colorectal cancer cells | Oncogene

The Role of Charge Density Coupled DNA Bending in Transcription Factor Sequence Binding Specificity: A Generic Mechanism for Indirect Readout | Journal of the American Chemical Society

Predicting transcription factor binding sites using DNA shape features based on shared hybrid deep learning architecture: Molecular Therapy - Nucleic Acids
Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain | PLOS Genetics

Systematic analysis of low-affinity transcription factor binding site clusters in vitro and in vivo establishes their functional relevance | Nature Communications
A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer | PLOS Genetics
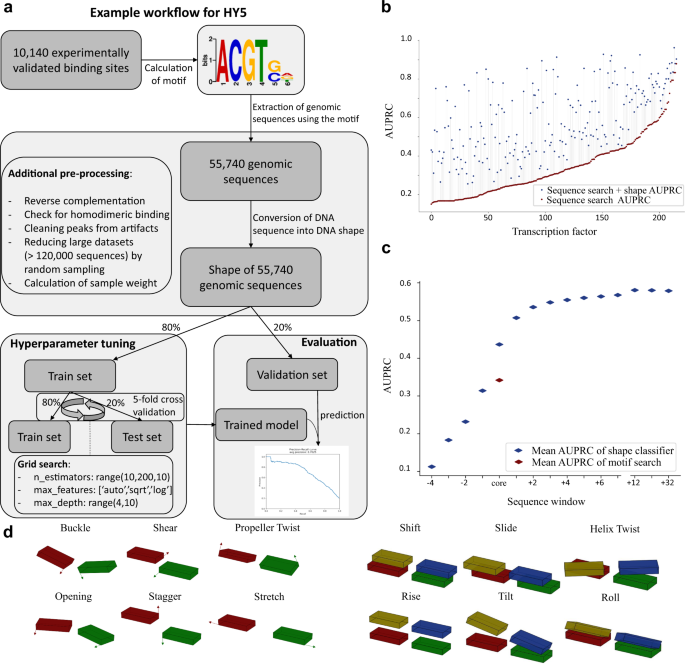
Local DNA shape is a general principle of transcription factor binding specificity in Arabidopsis thaliana | Nature Communications
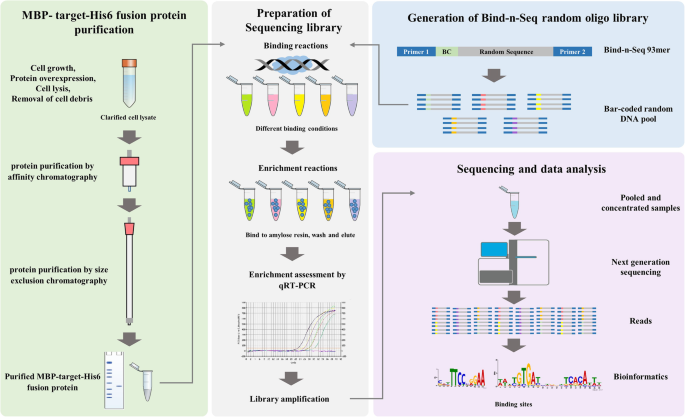
An improved bind-n-seq strategy to determine protein-DNA interactions validated using the bacterial transcriptional regulator YipR | BMC Microbiology | Full Text
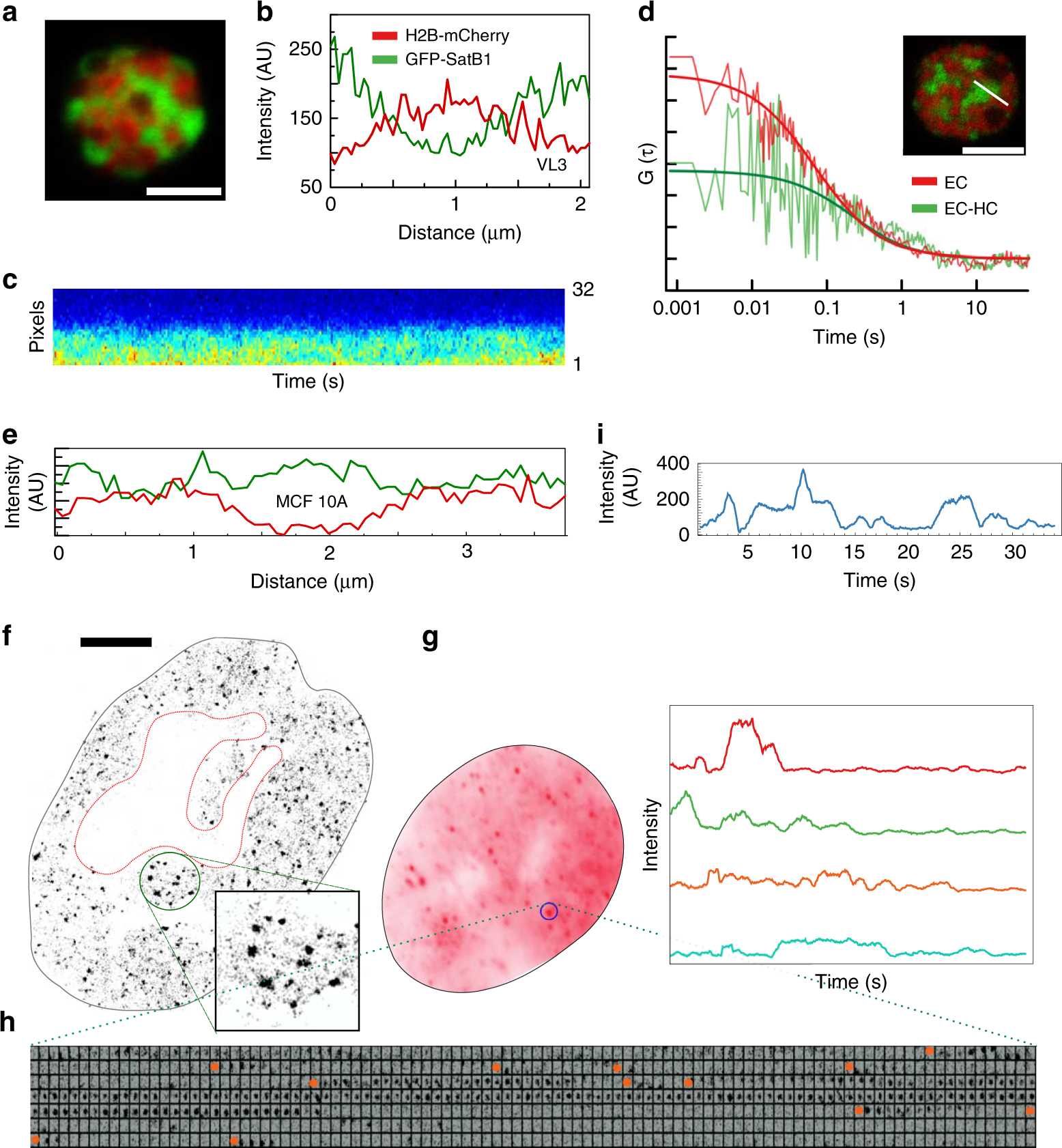
Satb1 integrates DNA binding site geometry and torsional stress to differentially target nucleosome-dense regions | Nature Communications

Coregulation of Transcription Factor Binding and Nucleosome Occupancy through DNA Features of Mammalian Enhancers - ScienceDirect






![PDF] Protein–DNA binding: complexities and multi-protein codes | Semantic Scholar PDF] Protein–DNA binding: complexities and multi-protein codes | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/da4fc83ebc798bbe5d356dd30cb2ae14fb01ae61/5-Figure1-1.png)


